DIFFICULT TEMPLATE SEQUENCING
Difficult Template sequencing is a proprietary Sanger protocol adept at tackling GC-rich regions, hairpins, and intricate secondary structures, ensuring a success rate more than double that of standard workflows. With GENEWIZ Difficult Template sequencing from Azenta Life Sciences, save at least 1 day in the sequencing process, and streamline your research.
Compatible with various DNA types including PCR products, plasmids, bacterial colonies, and glycerol stocks, our service eliminates the need for additional templates, providing unparalleled convenience and efficiency for your breakthrough projects.
Alternative Protocols for Difficult Templates
Template examples that may benefit from the alternative protocols include:
- GC-rich: A template of > 60% GC content or high GC-content concentrated in a small region
- Hairpin Structures: A sequence containing two inverted repeats, separated by at least three nucleotides
- Repeats: Certain di- or tri-nucleotide repeats
- Motifs causing band compressions: Mainly 5'-YGN1-2AR motifs, (Y: pyrimidine; R: purine; N: any base)
- Bacterial Artificial Chromosomes (BAC): Any construct larger than 50 kb.
Template Examples
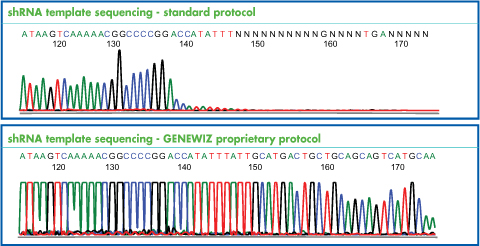
Case 1: Small Hairpin RNA (shRNA) Template Sequencing
The small hairpin RNA templates used in RNAi studies frequently present challenges in sequencing such as early termination or ambiguity, caused by the double-stranded or hairpin structures. Our proprietary RNAi Protocol produces superior results for our customers, saving time and cost. For a comparison, see the chromatogram below:
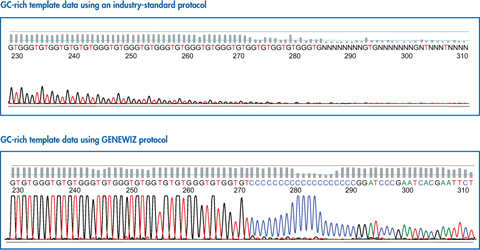
Case 2: Repeat / GC rich Template Sequencing
Repeat/GC-rich templates have special secondary structures and melting temperatures. DNA containing high GC content and hairpin structures are relatively more difficult to denature and very challenging to sequence.
Our proprietary Hairpin and GC-rich protocols produce superior results, with strong signals and uniform peak heights at the GC region.
BAC-end Sequencing for Constructs Greater than 50 kb
Large constructs such as Bacterial Artificial Chromosomes (BACs), cosmids, fosmids, and BACmids, pose a challenge to traditional Sanger sequencing protocols due to the size and interference of the non-target DNA in the template. GENEWIZ's proprietary BAC protocol to overcome the difficulty of sequencing directly from these large constructs can help deliver greater success and longer read lengths for large constructs compared to the standard protocol.
Please contact our Technical Support team for more information on BAC protocol's:
Email | Phone (1-877-GENEWIZ (436-3949), Ext. 2) | Live Chat
GENEWIZ provides our difficult template sequencing service (see above) using modified protocols, developed in-house, specifically for success with difficult templates. Years of use have confirmed that these alternative protocols deliver best-in-class results.
GENEWIZ provides individualized scientific consultation and customized solutions for your specific DNA sequencing needs, through our Technical Support team.
Our Technical Support team, staffed by multiple Ph.D. scientists with a friendly, problem-solving spirit, is available to advise and assist you at no charge Monday - Friday, 8AM - 8PM Eastern. GENEWIZ Technical Support also posts results on Saturdays, so research progress can continue forward over the weekend.

 Europe (English)
Europe (English)