Direct Colony Sequencing
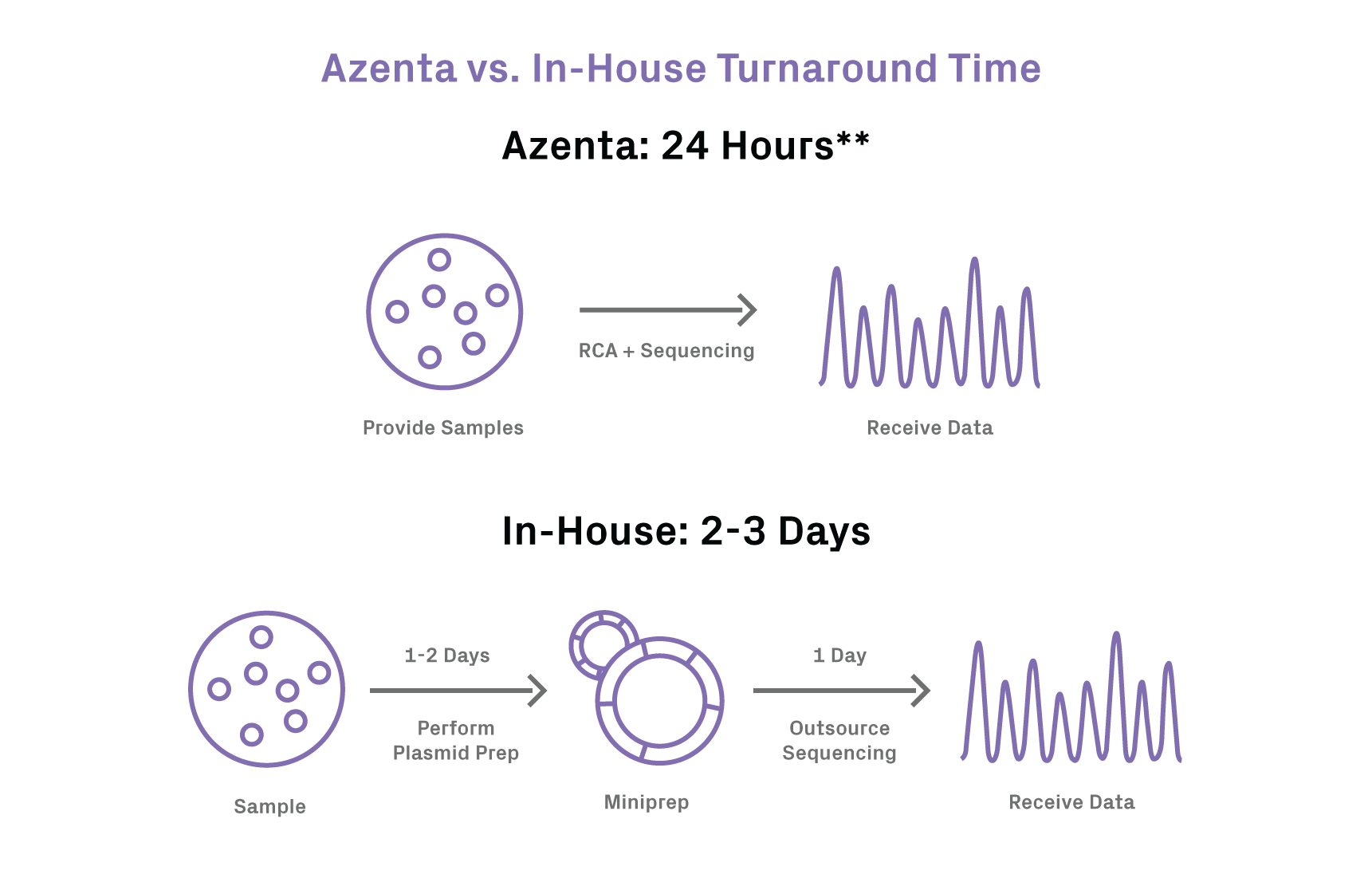
Direct Colony Sequencing services utilize rolling circle amplification (RCA) to enable Sanger sequencing of bacterial clone or phage sample templates without the need for plasmid preparation. RCA generates DNA for sequencing by random hexamer priming of circular templates. GENEWIZ RCA protocols can handle sequencing projects of any size and are optimized to produce high quality reads from both bacterial colonies and glycerol stocks as well as phage plaques and supernatants. Direct Colony Sequencing is also extremely effective when low amounts of DNA starting material are available (e.g. low copy plasmids).
What is colony sequencing?
Colony sequencing is a method in which plasmid DNA (pDNA) is extracted and amplified directly from microbial colonies, allowing for sequencing data to be produced without the need for plasmid purification. In addition to saving time by eliminating plasmid preparation steps, another key benefit of colony sequencing is its flexibility with the starting sample. Any size plasmid, with a high or low copy plasmid template, can be sequenced with direct colony sequencing. Direct colony sequencing is commonly used for screening complimentary DNA (cDNA) libraries and site-directed mutagenesis.
Rolling Circle Amplification Sequencing Protocol
Rolling Circle Amplification Sequencing
Rolling Circle Amplification was identified in natural systems as a DNA replication mechanism frequently employed in plasmid and viral propagation. When random hexamers are coupled with a strand displacing DNA polymerase, many concatenated template copies are generated with no need for thermocycling (Dean et al., 2001). The synthesized strands are displaced after one round of RCA and are free for additional rounds of primer binding. Exponential amplification is achievable if the RCA reaction contains primers complementary to both template strands
Dean, Frank B. et al. “Rapid Amplification of Plasmid and Phage DNA Using Phi29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification.” Genome Research 11.6 (2001): 1095–1099.
GENEWIZ Whole Genome Sequencing Workflow
-
After lysis, bacterial samples are combined with dNTPs, polymerase, and random hexamers. The hexamers anneal to the circular template, and replication initiates at many sites simultaneously.
-
Polymerase is utilized due to its high stability, processivity, and high fidelity. This allows prolonged strand displacement over multiple rounds of amplification.
-
As the newly synthesized strands are displaced, a cascade of priming events facilitates exponential amplification of the template.
-
Upon completion the sample is ready for Sanger sequencing.
Direct Colony Sequencing Features & Benefits
-
Time and cost savings: no need to grow culture and prepare DNA
-
High-quality data: >600 bases of Phred20 reads
-
Project design and consultation: Ph.D.-level customer support throughout the project
-
Fast turnaround: receive your Sanger data within 24 hours** of sample receipt at GENEWIZ
-
Online ordering: 24/7 online ordering and data retrieval system
-
GENEWIZ universal primers: select from a wide range of primers available at no additional cost
**24-hour turnaround time guaranteed for maximum of 96 samples; excludes phage samples
Rolling Circle Amplification Sequencing Applications
-
shRNA cloning and library screening; Eliminate plasmid preps and sequence colonies directly
-
DNA methylation analysis: Save time & obtain high-quality reads by cloning PCR products directly and sequencing the bacterial colonies
-
Antibody phage display library validation: Direct phage sequencing for faster screening and validation
Sample Submission Options
Sample Submission Options
| Glycerol Stock* | Send 1.5 ml tubes or 96-well plates; samples must be shipped on dry ice. Please label with media type. |
|---|---|
| Bacterial Colonies* | Array colonies in a grid on the agar plate and circle the colonies requiring sequencing. Numerically label each colony. Note that unnumbered plates will be processed as random pick. If using liquid culture, submit samples in 96-well plate with strip caps. Wrap agar plates in parafilm, and provide a cushion to protect against potential damage. |
| Phage Supernatant* | Please submit 50 ul of phage supernatant in 8-strip PCR tubes or 1.5 ml microcentrifuge tubes clearly labeled with sample name. |
| Phage Plaque | Circle and number the plaques you submit for sequencing. For transport, wrap agar plates in parafilm and provide cushion to protect against potential damage. |
*Please note: RCA protocol is intended for circular templates <10 kb; linear templates will not be amplified sufficiently for direct Sanger sequencing.
Longer circular templates, or circular templates carried in bacterial strains with functional endA may not be reliably amplified, contributing to variable results.
Please visit our Sample Submission Guidelines for bacteria and phage templates for additional details.
Technical Resources
-

Guide | Sanger Sequencing Quick Tips Guide
Learn from the Sanger experts how you can get the most of your DNA Sequencing. Whether you’re troubleshooting a tough sequencing reaction, perfecting PCR visualization, or learning to clean up and refine your PCR products, these resources are designed to help you get the best results.
- Volume 1: Producing Robust, Single-band PCR Product
- Volume 2: PCR Clean-up of Single-band PCR
- Volume 3: Salvaging Nonspecific PCR Products
- Volume 4: PCR Visualization by Gel Electrophoresis
- Volume 5: Troubleshooting a Bad Sequencing Reaction from a PCR Template
-

Blog | Troubleshooting DNA Templates with Sanger Sequencing
Sanger sequencing is highly effective for testing small targeted genomic regions and for validating results from NGS. However, as with all sequencing technologies, there are limitations and needs for troubleshooting. Read on to learn about the three basic troubleshooting steps we recommend to start with.
-

Webinar | From Base Pairs to Breakthroughs: Understanding Sanger and Oxford Nanopore Sequencing
While Sanger sequencing remains the gold standard for accurate DNA sequencing, advancements in sequencing technology, such as Oxford Nanopore Technology (ONT), have provided a cost-effective, long-read alternative for analysis. In this workshop, delve into the considerations of each technique and discover how to interpret their resulting sequencing data effectively so you can capture the benefits of both approaches for your specific needs.
-

Blog | Analyzing Sanger Sequencing Data
The output for Sanger sequencing is typically a chromatogram, also known as a trace or ab1 file, and a text-based sequence file. Although the latter may seem to hold all the relevant information—after all, the point of sequencing is to get a sequence—the former can’t be ignored. Here, we provide a guide to understanding Sanger sequencing data.
