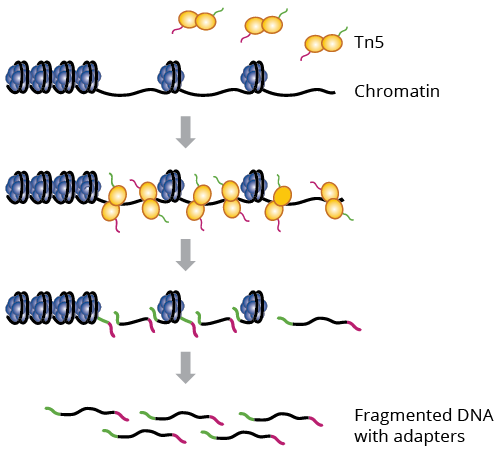
ATAC-Seq Workflow
1. Sample Preparation
Isolate and freeze cells with protocol provided by Azenta
2. Dead Cell Removal
If low viability, live cells are enriched using a magnetic bead-based approach
3. Nucleic Extraction & Library Preparation
Optimized ATAC assay produces robust signal from low inputs
4. Sequencing
Guaranteed data quality exceeds Illumina® benchmarks
5. Bioinformatics
Alignment, peak calling, and multiomic data integration

 Europe (English)
Europe (English)